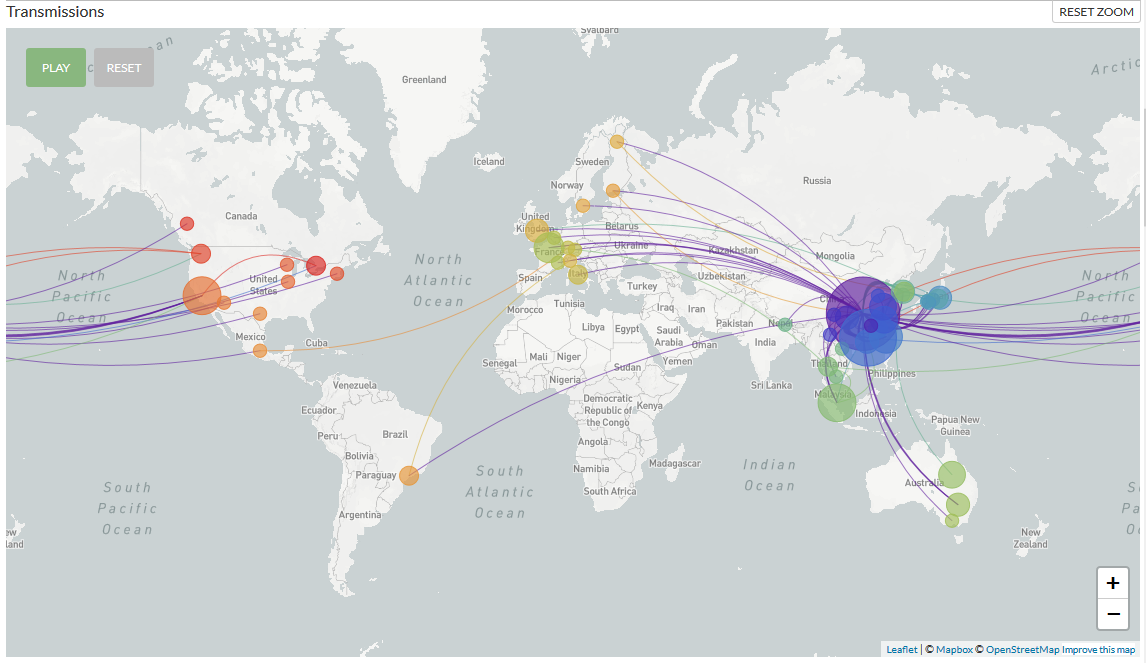
Latest Nextstrain COVID-19 situation report in English and in other languages
This phylogeny shows evolutionary relationships of HCoV-19 viruses from the ongoing novel coronavirus COVID-19 pandemic. All samples are still closely related with few mutations relative to a common ancestor, suggesting a shared common ancestor some time in Nov-Dec 2019. This indicates an initial human infection in Nov-Dec 2019 followed by sustained human-to-human transmission leading to sampled infections.
Site numbering and genome structure uses Wuhan-Hu-1/2019 as reference. The phylogeny is rooted relative to early samples from Wuhan. Temporal resolution assumes a nucleotide substitution rate of 5 × 10^-4 subs per site per year. Full details on bioinformatic processing can be found here.
Phylogenetic context of nCoV in SARS-related betacoronaviruses can be seen here.
We gratefully acknowledge the authors, originating and submitting laboratories of the genetic sequence and metadata made available through GISAID on which this research is based. A full listing of all originating and submitting laboratories is available below. An attribution table is available by clicking on "Download Data" at the bottom of the page and then clicking on "Strain Metadata" in the resulting dialog box.